全世界最新試管嬰兒新技術最新研究:次世代定序試管嬰兒 NGS
他們石破天驚發表在人類生殖期刋2014年10月21日,
一份使用NGS次世代定序應用在胚胎著床前基因診斷的試管嬰兒55位病人,研究當中55位病人平均年齡39.3歲,
他們檢測的胚胎都是囊胚期切片,切片切外胚層胚胎,第五天切片,
如果分裂得比較慢就第六天切片,甚至第七天才切片,
使用的方法是全基因放大,再使用次世代定序NGS,
並且用array CGH去做對比驗證,
使用的儀器是美國的Miseq探針,它是一種非常快速的時時間,由美國的illumina公司所生產,來算出染色體DNA的數量,
總共55位病人裡面分析了192個囊胚,
病人是使用次世代定序跟array CGH交叉分析,
在192個裡面有191個有訊號,其中有1個胚胎沒訊號,
總共測試了4608條染色體,
在這裡面有211條占4.6%是胚胎染色體非平衡性轉位,
在次世代定序裡面,它的準確性高達99.98%,也就是將近百分之百的準確性,
染色體正常的胚胎總共有50個胚胎植入到47個病人子宮,
也就是有些人植入一顆胚胎,有些人植入兩顆,
這47個裡面有34個懷孕,驗孕有,占72。34%,
但其中有4個驗孕有後來流掉了,8。5%
有30個持續懷孕63。8,
這30個應付生下31個試管寶寶,
繼續懷孕為63.8%,活產率63。8%,
值得一提的是:
在這個染色體裡面,
絕大部份是染色體異常的,因為191個裡面,真正染色體正常的只有74顆(38。7%),其中117 (61。25%)顆竟然是異常的,有74顆正常,
佔38.5%(74/191)而已,其中有117顆是染色體異常,佔61.5%
換句話說,在大自然的情況下,
染色體不正常是118顆vs比上染色體正常74顆,
也就是說,不正常的佔61.5%,正常的才佔38.5%,
這個比例可以讓病人和醫師充分了解;
其實我們在試管嬰兒已經是最好的囊胚的情況下,
仍然只有1/3是正常,2/3是不正常的,
這個是非常驚人的報告,
因為這是全世界第一個使用次世代定序NGS用之於臨床上的病人,
而締造了
64%的胚胎著床率,
63.8%懷孕率,
63。8%活產率,
不過無可諱言的是他們只植入1顆~2顆的胚胎,
那如果說是植入2顆~3顆胚胎呢?
不過話說回來,
經過次世代定序NGS的嚴格挑選,
在191個胚胎裡面,其實可以植入的也只有74個,
這74個如果分到47個病人,
一個人是1.57顆胚胎,
結論:
(1)一個病人進行試管嬰兒,能夠得到平均染色體正常的胚胎為1.4顆,
(2)植入一個囊胚期染色體正常為64%懷孕率,或64%著床率,
(3)大部分做出來的染色體,2/3為染色體異常的,佔61.5%,而不正常的佔38.5%
Hum Reprod. 2014 Oct 21. pii: deu277. [Epub ahead of print]
Application of next-generation sequencing technology for comprehensive aneuploidy screening of blastocysts in clinical preimplantation genetic screening cycles.
Abstract
STUDY QUESTION:
Can next-generation sequencing (NGS) techniques be used reliably for comprehensive aneuploidy screening of human embryos from patients undergoing IVF treatments, with the purpose of identifying and selecting chromosomally normal embryos for transfer?
SUMMARY ANSWER:
Extensive application of NGS in clinical preimplantation genetic screening (PGS) cycles demonstrates that this methodology is reliable, allowing identification and transfer of euploid embryos resulting in ongoing pregnancies.
WHAT IS KNOWN ALREADY:
The effectiveness of PGS is dependent upon the biology of the early embryo and the limitations of the technology. Fluorescence in situ hybridization, used to test for a few chromosomes, has largely been superseded by microarray techniques that test all 24 chromosomes. Array comparative genomic hybridization (array-CGH) has been demonstrated to be an accurate PGS method and has become the de facto gold standard, but new techniques, such as NGS, continue to emerge.
STUDY DESIGN, SIZE, DURATION:
The study consisted of a prospective trial involving a double blind parallel evaluation, with both NGS and array-CGH techniques, of 192 blastocysts obtained from 55 consecutive clinical PGS cycles undertaken during the period of September to October 2013. Consistency of NGS-based aneuploidy detection was assessed by matching the results obtained with array-CGH-based diagnoses. Primary outcome measure was accuracy of the chromosomal analysis; secondary outcome measures were clinical outcomes.
PARTICIPANTS/MATERIALS, SETTINGS, METHODS:
Fifty-five patients (median age 39.3 years, range 32-46) undergoingPGS were enrolled in the study. All embryos were cultured to blastocyst stage; trophectoderm biopsy was performed on Day 5 of development or Day 6/7 for slower growing embryos. The method involved whole genome amplification followed by both NGS and array-CGH. The MiSeq® control software, real-time analysis and reporter performed on-board primary and secondary bioinformatics analysis. Copy number variation analysis was accomplished with BlueFuse Multi software.
MAIN RESULTS AND THE ROLE OF CHANCE:
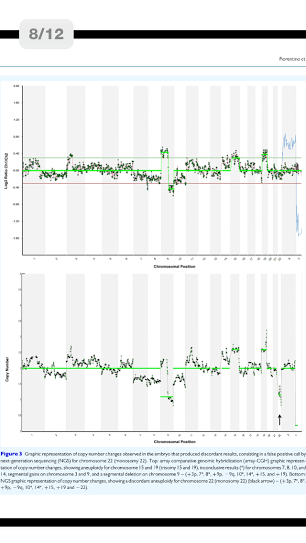
A total of 192 blastocysts were blindly evaluated with the NGS-based protocol. Paired comparison between NGS and array-CGH from individual embryos showed concordant results in 191/192 (99.5%) of the blastocysts tested. In total 4608 chromosomes were assessed, 211 (4.6%) of which carried a copy number imbalance. NGS specificity for aneuploidy calling (consistency of chromosome copy number assignment) was 99.98% (4333/4334; 95% confidence interval [95% CI]: 99.87-100) with a sensitivity of 100% (211/211, 95% CI: 99.25-100). Despite one discordant result, NGS specificity and sensitivity for aneuploid embryo calling (24-chromosome diagnosis consistency) were both 100% since the discordant sample presented several other aneuploidies. Clinical application of the NGS-based approach revealed 74/192 (38.5%) euploid blastocysts. Following transfer of 50 embryos in 47 women, 34 women had positive hCG levels: 30 pregnancies continued, confirmed by at least one fetal sac and heart beat (63.8% clinical pregnancy rate/embryo transfer), 3 were biochemical and 1 miscarried. A total of 32 embryos implanted and led to the presence of a fetal sac (64.0% implantation rate). All pregnancies went to term resulting in the birth of 31 healthy babies.
LIMITATION, REASON FOR CAUTION:
Although clinical results reported high pregnancy outcomes following transfer of screened embryos, further data and broad-based clinical application are required to better define the role of NGS in PGS. Before recommending widespread application, a randomized controlled trial confirming its clinical effectiveness is advisable.
WIDER IMPLICATION OF THE FINDING:
This is the first study reporting extensive application of NGS-based comprehensive aneuploidy screening on embryos at blastocyst stage in a clinical setting versus array-CGH as test of reference. NGS has demonstrated a reliable methodology, with the potential to improve chromosomal diagnosis on embryos especially in terms of high-throughput, automation and ability to detect aneuploidy. NGS methodology may represent a valuable alternative to the other comprehensive aneuploidy screening techniques currently available.
STUDY FUNDING/COMPETING INTERESTS:
No external funding was sought for this study. Drs F.K. and C.-E.M. are full-time employees of Illumina, Inc., which provided NGS library and sequencing reagents for the study. All other authors have no conflicts to declare.
TRIAL REGISTRATION NUMBER:
Not applicable.
© The Author 2014. Published by Oxford University Press on behalf of the European Society of Human Reproduction and Embryology. All rights reserved. For Permissions, please email:journals.permissions@oup.com.
KEYWORDS:
array-comparative genomic hybridization; clinical outcomes; comprehensive chromosome screening; next-generation sequencing; preimplantation genetic screening